Natural Language Processing of German texts - Part 2: Using LSTM neural-networks to predict ratings
In the first part of this series, we implemented a complete machine-learning workflow for binary text classification. Using a unique German data set, we achieved decent results in predicting doctor ratings from patients’ text comments.
Here, we will improve on our results by applying more advanced modeling techniques. Namely, we will use Tensorflow / Keras to implement Long Short Term Memory (LSTM) neural networks and use self trained and pre trained embeddings (i.e. FastText). For that, we will adapt the previous work-flow and focus on feature creation and modeling:
For the feature creation, we will use embeddings. These will be created in Keras. Similarly, for the modeling we will use Keras to implement our LSTM neural network for the classification task. Cleaning, pre-processing and evaluation can easily be adopted from our original work-flow. In the end, we will compare our results to the baseline we established in part 1. In the following notebook, we will go through this process step by step.
You can download this notebook or follow along in an interactive version of it on and
.
Setup / Data set / cleaning / pre processing
As this is not the focus of this post, we will go through these steps rather quickly. If you need a refresher, check out the first post again.
We’ll be using the same data as before. You can take a look at it on data.world or directly download it from here.
While the first part heavily relied on scikit-learn, here we will use Tensorflow and more specifically Keras for implementing our model. Neural Networks are much more computationally demanding compared to other machine-learning methods. They can benefit from using a GPU and even more from using a TPU for computation. Running them on a CPU will usually be frustrating as it can take ages with larger data sets. Luckily, the notebooks on Google Colab offer free TPU usage. If you want to replicate this post, your best bet is to make use of this.
import os
import random
import nltk
import re
import pickle
import pandas as pd
import numpy as np
import tensorflow as tf
import seaborn as sns
from datetime import datetime
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Input, LSTM, Bidirectional, Dense
from tensorflow.keras.layers import Embedding, Flatten
from tensorflow.keras.layers import MaxPooling1D, Dropout, Activation, Conv1D
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing import sequence
from tensorflow.keras.callbacks import EarlyStopping, ModelCheckpoint
from tensorflow.keras.models import load_model
from sklearn import metrics
from sklearn.model_selection import train_test_split
from nltk.stem.snowball import SnowballStemmer
from nltk.tokenize import word_tokenize
from nltk.corpus import stopwords
import matplotlib.pyplot as plt
import warnings
pd.options.display.max_colwidth = 6000
pd.options.display.max_rows = 400
np.set_printoptions(suppress=True)
warnings.filterwarnings("ignore")
print(tf.__version__)
2.2.0
# Needed on Google Colab
if os.environ.get('COLAB_GPU', False):
!pip install -U holoviews hvplot panel==0.8.1
Executing this on Colab will make sure that our model runs on a TPU if available and falls back to GPU / CPU otherwise:
# Try to run on TPU
# Detect hardware, return appropriate distribution strategy
try:
tpu = tf.distribute.cluster_resolver.TPUClusterResolver() # TPU detection
print("Running on TPU ", tpu.cluster_spec().as_dict()["worker"])
except ValueError:
tpu = None
if tpu:
tf.config.experimental_connect_to_cluster(tpu)
tf.tpu.experimental.initialize_tpu_system(tpu)
strategy = tf.distribute.experimental.TPUStrategy(tpu)
else:
strategy = tf.distribute.get_strategy()
print("REPLICAS: ", strategy.num_replicas_in_sync)
Running on TPU ['10.2.239.34:8470']
INFO:tensorflow:Initializing the TPU system: grpc://10.2.239.34:8470
INFO:tensorflow:Clearing out eager caches
INFO:tensorflow:Finished initializing TPU system.
INFO:tensorflow:Found TPU system:
INFO:tensorflow:*** Num TPU Cores: 8
INFO:tensorflow:*** Num TPU Workers: 1
INFO:tensorflow:*** Num TPU Cores Per Worker: 8
INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:localhost/replica:0/task:0/device:CPU:0, CPU, 0, 0)
INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:localhost/replica:0/task:0/device:XLA_CPU:0, XLA_CPU, 0, 0)
INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:CPU:0, CPU, 0, 0)
INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:0, TPU, 0, 0)
INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:1, TPU, 0, 0)
INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:2, TPU, 0, 0)
INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:3, TPU, 0, 0)
INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:4, TPU, 0, 0)
INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:5, TPU, 0, 0)
INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:6, TPU, 0, 0)
INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:7, TPU, 0, 0)
INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU_SYSTEM:0, TPU_SYSTEM, 0, 0)
INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:XLA_CPU:0, XLA_CPU, 0, 0)
REPLICAS: 8
In our case, we are running on 8 TPUs cores. This will give us an immense speed up!
As before, we first download and extract our data:
# store current path and download data there
CURR_PATH = !pwd
!wget -O reviews.zip https://query.data.world/s/v5xl53bs2rgq476vqy7cg7xx2db55y
!unzip reviews.zip
--2020-06-06 17:33:00-- https://query.data.world/s/v5xl53bs2rgq476vqy7cg7xx2db55y
Resolving query.data.world (query.data.world)... 35.174.192.118, 34.231.236.159, 54.165.184.72
Connecting to query.data.world (query.data.world)|35.174.192.118|:443... connected.
HTTP request sent, awaiting response... 301 Moved Permanently
Location: https://download.data.world/file_download/mc51/german-language-reviews-of-doctors-by-patients/german_doctor_reviews.zip?auth=eyJhbGciOiJIUzUxMiJ9.eyJzdWIiOiJwcm9kLXVzZXItY2xpZW50Om1jNTEiLCJpc3MiOiJhZ2VudDptYzUxOjoxMTRmMjJkZi1jMTkxLTRlNGYtYmNjZC01NTZhMzc0M2ZiOTkiLCJpYXQiOjE1ODI5OTUwMDEsInJvbGUiOlsidXNlciIsInVzZXJfYXBpX2FkbWluIiwidXNlcl9hcGlfcmVhZCIsInVzZXJfYXBpX3dyaXRlIl0sImdlbmVyYWwtcHVycG9zZSI6ZmFsc2UsInVybCI6IjJkNTVlNDU3YzQ3ZGI5MGUwNzMxODAwMTdhZjk5YWY0ODc3ZjYwYTAifQ.jcIyJu6pFRC6R8zmoB0fU4s8pyKO4SImC6kKoxFVCIhzok5_dWYTzncgQ8WU4Uw3NSGxI4oh7YpZFsyfl3H-qg [following]
--2020-06-06 17:33:00-- https://download.data.world/file_download/mc51/german-language-reviews-of-doctors-by-patients/german_doctor_reviews.zip?auth=eyJhbGciOiJIUzUxMiJ9.eyJzdWIiOiJwcm9kLXVzZXItY2xpZW50Om1jNTEiLCJpc3MiOiJhZ2VudDptYzUxOjoxMTRmMjJkZi1jMTkxLTRlNGYtYmNjZC01NTZhMzc0M2ZiOTkiLCJpYXQiOjE1ODI5OTUwMDEsInJvbGUiOlsidXNlciIsInVzZXJfYXBpX2FkbWluIiwidXNlcl9hcGlfcmVhZCIsInVzZXJfYXBpX3dyaXRlIl0sImdlbmVyYWwtcHVycG9zZSI6ZmFsc2UsInVybCI6IjJkNTVlNDU3YzQ3ZGI5MGUwNzMxODAwMTdhZjk5YWY0ODc3ZjYwYTAifQ.jcIyJu6pFRC6R8zmoB0fU4s8pyKO4SImC6kKoxFVCIhzok5_dWYTzncgQ8WU4Uw3NSGxI4oh7YpZFsyfl3H-qg
Resolving download.data.world (download.data.world)... 34.231.236.159, 35.174.192.118, 54.165.184.72
Connecting to download.data.world (download.data.world)|34.231.236.159|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: unspecified [application/zip]
Saving to: ‘reviews.zip’
reviews.zip [ <=> ] 42.94M 72.9MB/s in 0.6s
2020-06-06 17:33:01 (72.9 MB/s) - ‘reviews.zip’ saved [45022322]
Archive: reviews.zip
inflating: german_doctor_reviews.csv
# On Colab mount our google drive for storing data
if os.environ.get('COLAB_GPU', False):
from google.colab import drive
drive.mount("/drive")
Here we set the parameters that we will use for embedding and the neural network. The batch size will depend on the number of available cores. We’ll look at the other parameters later on.
# PARAMETERS
PATH_DATA = CURR_PATH[0]
PATH_MODELS = PATH_DATA + "models/"
PATH_CHECKPOINTS = PATH_MODELS + "checkpoints/"
# Text Vectors
MAX_FEATURES = 30000
EMBED_DIM = 300
MAXLEN = 400
# Convolution
KERNEL_SIZE = 5
FILTERS = 64
POOL_SIZE = 4
# LSTM
LSTM_OUTPUT_SIZE = 100
# Training
BATCH_SIZE = 32 * strategy.num_replicas_in_sync
EPOCHS = 10
Let’s load the data set and create our target variable. Positive ratings (one or two) will be considered as good and negative ones (five or six) as negative. As before, we ignore neutral ratings:
# read data from csv
data = pd.read_csv(PATH_DATA + "/german_doctor_reviews.csv")
# Create binary grade, class 1-2 or 5-6 = good or bad
data["grade_bad"] = 0
data.loc[data["rating"] >= 3, "grade_bad"] = np.nan
data.loc[data["rating"] >= 5, "grade_bad"] = 1
data.head(2)
| comment | rating | grade_bad | |
|---|---|---|---|
| 0 | Ich bin franzose und bin seit ein paar Wochen in muenchen. Ich hatte Zahn Schmerzen und mein Kollegue hat mir Dr mainka empfohlen. Ich habe schnell ein Termin bekommen, das Team war nett und meine schmerzen sind weg!! Ich bin als Angst Patient sehr zurieden!! | 2.0 | 0.0 |
| 1 | Dieser Arzt ist das unmöglichste was mir in meinem Leben je begegnet ist,er ist unfreundlich ,sehr herablassend und medizinisch unkompetent.Nach seiner Diagnose bin ich zu einem anderen Hautarzt gegangen der mich ordentlich behandelt hat und mir auch half.Meine Beschweerden hatten einen völlig anderen Grund.<br />\nNach seiner " Behandlung " und Diagnose ,waren seine letzten Worte .....und tschüss.Alles inerhalb von ca 5 Minuten. | 6.0 | 1.0 |
Our cleaning and pre-processing strategy is very similar to the first part. However, when using frequency based methods for feature creation, it is paramount to “distill” the texts. You want to remove irrelevant text bits so that only informative content remains. Also, when using rather simple methods for prediction, the text should be simple as well. Those methods will not be able to pick up on sophisticated associations. In contrast, neural networks are able to pick up on more subtle relationships and can learn to automatically ignore uninformative parts. Following, they require less manipulation of the input texts. Hence, we will skip some pre processing steps used before, i.e. there will be no stopword and punctuation removal and also no stemming. However, keep in mind that there is no absolute truth. You will need to experiment quite a bit to find out which pre-processing strategy works best in your specific case!
nltk.download("stopwords")
nltk.download("punkt")
stemmer = SnowballStemmer("german")
stop_words = set(stopwords.words("german"))
def clean_text(text, for_embedding=False):
"""
- remove any html tags (< /br> often found)
- Keep only ASCII + European Chars and whitespace, no digits
- remove single letter chars
- convert all whitespaces (tabs etc.) to single wspace
if not for embedding (but e.g. tdf-idf):
- all lowercase
- remove stopwords, punctuation and stemm
"""
RE_WSPACE = re.compile(r"\s+", re.IGNORECASE)
RE_TAGS = re.compile(r"<[^>]+>")
RE_ASCII = re.compile(r"[^A-Za-zÀ-ž ]", re.IGNORECASE)
RE_SINGLECHAR = re.compile(r"\b[A-Za-zÀ-ž]\b", re.IGNORECASE)
if for_embedding:
# Keep punctuation
RE_ASCII = re.compile(r"[^A-Za-zÀ-ž,.!? ]", re.IGNORECASE)
RE_SINGLECHAR = re.compile(r"\b[A-Za-zÀ-ž,.!?]\b", re.IGNORECASE)
text = re.sub(RE_TAGS, " ", text)
text = re.sub(RE_ASCII, " ", text)
text = re.sub(RE_SINGLECHAR, " ", text)
text = re.sub(RE_WSPACE, " ", text)
word_tokens = word_tokenize(text)
words_tokens_lower = [word.lower() for word in word_tokens]
if for_embedding:
# no stemming, lowering and punctuation / stop words removal
words_filtered = word_tokens
else:
words_filtered = [
stemmer.stem(word) for word in words_tokens_lower if word not in stop_words
]
text_clean = " ".join(words_filtered)
return text_clean
[nltk_data] Downloading package stopwords to /root/nltk_data...
[nltk_data] Unzipping corpora/stopwords.zip.
[nltk_data] Downloading package punkt to /root/nltk_data...
[nltk_data] Unzipping tokenizers/punkt.zip.
Now, we can we apply this pre-processing and cleaning to our original data:
%%time
# Clean Comments
data["comment_clean"] = data.loc[data["comment"].str.len() > 20, "comment"]
data["comment_clean"] = data["comment_clean"].map(
lambda x: clean_text(x, for_embedding=True) if isinstance(x, str) else x
)
CPU times: user 3min 46s, sys: 219 ms, total: 3min 46s
Wall time: 3min 46s
This is how the final comments will look like:
# Drop Missing
data = data.dropna(axis="index", subset=["grade_bad", "comment_clean"]).reset_index(
drop=True
)
data.head(2)
| comment | rating | grade_bad | comment_clean | |
|---|---|---|---|---|
| 0 | Ich bin franzose und bin seit ein paar Wochen in muenchen. Ich hatte Zahn Schmerzen und mein Kollegue hat mir Dr mainka empfohlen. Ich habe schnell ein Termin bekommen, das Team war nett und meine schmerzen sind weg!! Ich bin als Angst Patient sehr zurieden!! | 2.0 | 0.0 | Ich bin franzose und bin seit ein paar Wochen in muenchen . Ich hatte Zahn Schmerzen und mein Kollegue hat mir Dr mainka empfohlen . Ich habe schnell ein Termin bekommen , das Team war nett und meine schmerzen sind weg ! ! Ich bin als Angst Patient sehr zurieden ! ! |
| 1 | Dieser Arzt ist das unmöglichste was mir in meinem Leben je begegnet ist,er ist unfreundlich ,sehr herablassend und medizinisch unkompetent.Nach seiner Diagnose bin ich zu einem anderen Hautarzt gegangen der mich ordentlich behandelt hat und mir auch half.Meine Beschweerden hatten einen völlig anderen Grund.<br />\nNach seiner " Behandlung " und Diagnose ,waren seine letzten Worte .....und tschüss.Alles inerhalb von ca 5 Minuten. | 6.0 | 1.0 | Dieser Arzt ist das unmöglichste was mir in meinem Leben je begegnet ist er ist unfreundlich , sehr herablassend und medizinisch unkompetent Nach seiner Diagnose bin ich zu einem anderen Hautarzt gegangen der mich ordentlich behandelt hat und mir auch half Meine Beschweerden hatten einen völlig anderen Grund . Nach seiner Behandlung und Diagnose , waren seine letzten Worte ... ..und tschüss Alles inerhalb von ca Minuten . |
As before, we split our data into a training and testing set for cross validation:
# Sample data for cross validation
train, test = train_test_split(data, random_state=1, test_size=0.25, shuffle=True)
X_train = np.array(train["comment_clean"])
Y_train = np.array(train["grade_bad"]).reshape((-1, 1))
X_test = np.array(test["comment_clean"])
Y_test = np.array(test["grade_bad"]).reshape((-1, 1))
print(X_train.shape)
print(X_test.shape)
(253782,)
(84595,)
Feature Creation
When dealing with text data, we need to convert text to a numeric representation first. For that, we use the Tokenizer function in Keras. After splitting the text into tokens (i.e. words or punctuation) the function assigns each unique token a number, i.e. it builds a token <-> integer dictionary. Using this, each comment is transformed to a vector of integers in which each element represents a token. Here, we also transform all tokens to lowercase and limit the maximum number of used unique tokens to MAX_FEATURES:
%%time
# create numerical vector representation of comments
# comment to list of indices representing words in dict
tokenizer = Tokenizer(lower=True, split=" ", num_words=MAX_FEATURES)
tokenizer.fit_on_texts(X_train)
X_train_vec = tokenizer.texts_to_sequences(X_train)
X_test_vec = tokenizer.texts_to_sequences(X_test)
MAXLEN = max([len(x) for x in X_train_vec])
print(f"Max vector length: {MAXLEN}")
Max vector length: 355
CPU times: user 38.3 s, sys: 101 ms, total: 38.4 s
Wall time: 38.4 s
Next, we make sure that our vectors have a fixed length equal to the maximal comment length. Shorter vectors will be padded with zeros:
# pad with zeros for same vector length
X_train_vec = sequence.pad_sequences(X_train_vec, maxlen=MAXLEN, padding="post")
X_test_vec = sequence.pad_sequences(X_test_vec, maxlen=MAXLEN, padding="post")
Now, we take a look at the results of the transformation:
tmp = train[0:1].copy()
tmp["vector"] = list(X_train_vec[0:1])
tmp
| comment | rating | grade_bad | comment_clean | vector | |
|---|---|---|---|---|---|
| 44011 | Aufgrund der vielen guten Bewertungen bin auch ich heute als "Notfall" bei Dr. Böhme gelandet. Habe am gleichen Tag auch einen Termin bekommen, die Online-Abwicklung war absolut perfekt!!<br />\r\nEr hat aufmerksam zugehört, als erster entdeckt, daß mein Herz etwas anders liegt als sonst üblich und ging auf meine mysteriösen Symptome mit Ruhe und freundlichem Interesse ein. Jetzt startet erst mal die Untersuchungsreihe, dann geht es um den weiteren Ablauf. Bis jetzt bin ich jedenfalls ausgesprochen angetan. Danke! | 1.0 | 0.0 | Aufgrund der vielen guten Bewertungen bin auch ich heute als Notfall bei Dr. Böhme gelandet . Habe am gleichen Tag auch einen Termin bekommen , die Online Abwicklung war absolut perfekt ! ! Er hat aufmerksam zugehört , als erster entdeckt , daß mein Herz etwas anders liegt als sonst üblich und ging auf meine mysteriösen Symptome mit Ruhe und freundlichem Interesse ein . Jetzt startet erst mal die Untersuchungsreihe , dann geht es um den weiteren Ablauf . Bis jetzt bin ich jedenfalls ausgesprochen angetan . Danke ! | [306, 5, 123, 176, 396, 23, 10, 2, 206, 40, 520, 15, 7, 5627, 889, 30, 86, 806, 207, 10, 43, 52, 154, 3, 906, 4077, 19, 166, 372, 26, 17, 400, 1004, 40, 858, 1662, 572, 96, 780, 140, 573, 578, 40, 559, 2132, 1, 247, 32, 50, 807, 14, 358, 1, 2230, 924, 21, 152, 19212, 236, 83, 3, 85, 120, 28, 82, 36, 488, 1097, 134, 152, 23, 2, 1316, 611, 2513, 185, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, ...] |
print(
f"The comment is transformed to a vector whose first element is \"{tmp['vector'].iloc[0][0]}\". This integer translates to: \"{tokenizer.index_word[tmp['vector'].iloc[0][0]]}\" which is the token representing the original word."
)
The comment is transformed to a vector whose first element is "306". This integer translates to: "aufgrund" which is the token representing the original word.
Defining the predictive model
So far, we hardly needed to adapt the previous work flow. In the following section this changes as we introduce a new modeling approach.
We start by defining our neural network layer by layer. First, we use an embedding layer. Its aim is to learn a dense vector representation for each token which maximizes the objective function of the network. These dense representations are in contrast to the sparse representations we got from using frequency based methods in part 1. One advantage is that neural networks can handle them much better as input features. Another one being that these vectors have helpful properties. For example, a model might assign similar vectors to words holding a similar meaning or those that have a comparable importance for the task at hand.
On top of the embedding layer we stack a dropout layer. This is supposed to reduce overfitting by randomly dropping nodes of the network while training.
Next, we add a convolutional layer. This might sound familiar from an image recognition context but has also found its way into NLP. By passing filters over it, this layer calculates a higher dimensionality of the data. Doing this, it can detect the most prominent patterns in the data while reducing the computational demand.
The pooling layer further reduces the dimensionality of the data and helps extract the most dominant patterns. Its side effect is that it helps with overfitting as well.
Next, we add the Long Short Term Memory (LSTM) layer. LSTM is a form of Recurrent Neural Network (RNN). RNNs have been terrfic in solving all kinds of problems by adding the ability to persist information over longer input sequences to traditional networks. Thus, they can take context into consideration which beautifully fits the demands of text understanding. In addition to that, LSTMs enable models to even take long term dependencies into account. Why is this helpful? Because to whom is refered to in the current sentence might depend on who was refered to in the previous one. In language context is not always immediate.
The model is completed using a dense layer which reduces the output to be either zero or one, giving us our class prediction.
# Define NN architecture
with strategy.scope():
model = Sequential()
model.add(
Embedding(input_dim=MAX_FEATURES, output_dim=EMBED_DIM, input_length=MAXLEN)
)
model.add(Dropout(0.3))
model.add(
Conv1D(FILTERS, KERNEL_SIZE, padding="valid", activation="relu", strides=1)
)
model.add(MaxPooling1D(pool_size=POOL_SIZE))
model.add(LSTM(LSTM_OUTPUT_SIZE))
model.add(Dense(1, activation="sigmoid"))
model.compile(
loss="binary_crossentropy",
optimizer=tf.keras.optimizers.RMSprop(),
metrics=["accuracy"],
)
Now, our model architecture looks like this:
model.summary()
Model: "sequential"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
embedding (Embedding) (None, 357, 300) 9000000
_________________________________________________________________
dropout (Dropout) (None, 357, 300) 0
_________________________________________________________________
conv1d (Conv1D) (None, 353, 64) 96064
_________________________________________________________________
max_pooling1d (MaxPooling1D) (None, 88, 64) 0
_________________________________________________________________
lstm (LSTM) (None, 100) 66000
_________________________________________________________________
dense (Dense) (None, 1) 101
=================================================================
Total params: 9,162,165
Trainable params: 9,162,165
Non-trainable params: 0
_________________________________________________________________
Here, we define a callback function that is getting called during training. It does two things. First, it stops the training if there is no more improvement. Second, it saves model checkpoints after each iteration (epoch):
# Stop training when validation acc starts dropping
# Save checkpoint of model each period
now = datetime.now().strftime("%Y-%m-%d_%H%M")
# Create callbacks
callbacks = [
EarlyStopping(monitor="val_loss", verbose=1, patience=2),
ModelCheckpoint(
PATH_CHECKPOINTS + now + "_Model_{epoch:02d}_{val_loss:.4f}.h5",
monitor="val_loss",
save_best_only=True,
verbose=1,
),
]
Finally, we train our model:
%%time
# Fit the model
steps_per_epoch = int(np.floor((len(X_train_vec) / BATCH_SIZE)))
print(
f"Model Params.\nbatch_size: {BATCH_SIZE}\nEpochs: {EPOCHS}\n"
f"Step p. Epoch: {steps_per_epoch}\n"
)
hist = model.fit(
X_train_vec,
Y_train,
batch_size=BATCH_SIZE,
epochs=EPOCHS,
steps_per_epoch=steps_per_epoch,
callbacks=callbacks,
validation_data=(X_test_vec, Y_test),
)
Model Params.
batch_size: 256
Epochs: 10
Step p. Epoch: 991
Epoch 1/10
991/991 [==============================] - ETA: 0s - loss: 0.2305 - accuracy: 0.9204
Epoch 00001: val_loss improved from inf to 0.13770, saving model to /drive/My Drive/tmp/2020-06-06_1748_Model_01_0.1377.h5
991/991 [==============================] - 28s 28ms/step - loss: 0.2305 - accuracy: 0.9204 - val_loss: 0.1377 - val_accuracy: 0.9454
Epoch 2/10
991/991 [==============================] - ETA: 0s - loss: 0.0862 - accuracy: 0.9662
Epoch 00002: val_loss improved from 0.13770 to 0.07425, saving model to /drive/My Drive/tmp/2020-06-06_1748_Model_02_0.0743.h5
991/991 [==============================] - 26s 26ms/step - loss: 0.0862 - accuracy: 0.9662 - val_loss: 0.0743 - val_accuracy: 0.9708
Epoch 3/10
991/991 [==============================] - ETA: 0s - loss: 0.0653 - accuracy: 0.9751
Epoch 00003: val_loss improved from 0.07425 to 0.06929, saving model to /drive/My Drive/tmp/2020-06-06_1748_Model_03_0.0693.h5
991/991 [==============================] - 26s 26ms/step - loss: 0.0653 - accuracy: 0.9751 - val_loss: 0.0693 - val_accuracy: 0.9722
Epoch 4/10
990/991 [============================>.] - ETA: 0s - loss: 0.0569 - accuracy: 0.9784
Epoch 00004: val_loss did not improve from 0.06929
991/991 [==============================] - 25s 25ms/step - loss: 0.0569 - accuracy: 0.9784 - val_loss: 0.0698 - val_accuracy: 0.9737
Epoch 5/10
991/991 [==============================] - ETA: 0s - loss: 0.0506 - accuracy: 0.9815
Epoch 00005: val_loss improved from 0.06929 to 0.06767, saving model to /drive/My Drive/tmp/2020-06-06_1748_Model_05_0.0677.h5
991/991 [==============================] - 25s 25ms/step - loss: 0.0506 - accuracy: 0.9815 - val_loss: 0.0677 - val_accuracy: 0.9734
Epoch 6/10
990/991 [============================>.] - ETA: 0s - loss: 0.0454 - accuracy: 0.9836
Epoch 00006: val_loss did not improve from 0.06767
991/991 [==============================] - 25s 25ms/step - loss: 0.0454 - accuracy: 0.9837 - val_loss: 0.0746 - val_accuracy: 0.9726
Epoch 7/10
991/991 [==============================] - ETA: 0s - loss: 0.0408 - accuracy: 0.9852
Epoch 00007: val_loss did not improve from 0.06767
991/991 [==============================] - 25s 25ms/step - loss: 0.0408 - accuracy: 0.9852 - val_loss: 0.0720 - val_accuracy: 0.9732
Epoch 00007: early stopping
CPU times: user 31.3 s, sys: 6.82 s, total: 38.1 s
Wall time: 3min 12s
After five epochs the training reaches its best performance. The validation accuracy is around 0.97. We can depict the training and test loss of the model, to get a sense for how well and fast our model has been learning:
loss = pd.DataFrame(
{"train loss": hist.history["loss"], "test loss": hist.history["val_loss"]}
).melt()
loss["epoch"] = loss.groupby("variable").cumcount() + 1
sns.lineplot(x="epoch", y="value", hue="variable", data=loss).set(
title="Model loss", ylabel=""
)
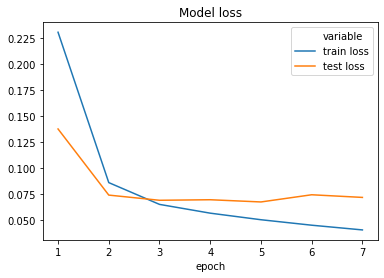
While the train loss keeps decreasing through all epochs, the test score stops dropping (and even slightly increases) after epoch five. Hence, it is sensible to stop the training here as going on would only lead to overfitting our model.
A look at learned embeddings
Following, we take a look at the embeddings that our network has learned. The embedding vectors correspond to the weights of the embedding layer. We use them to get the embeddings for the 10k most used words in the comments:
# get trained embeddings
embeddings = model.layers[0].get_weights()[0]
# get token <-> integer dictionary
word_index = tokenizer.word_index.items()
# for each word in dict get embedding
words_embeddings = {w: embeddings[idx] for w, idx in word_index if idx < 10000}
# show embedding vector for the word
words_embeddings.get("arzt")
array([ 0.05396529, -0.08387542, -0.19157657, -0.03766663, -0.01097831,
-0.00982297, -0.0831705 , -0.06066611, -0.1817672 , -0.03342694,
-0.00319161, -0.09381639, -0.11026296, 0.02463322, 0.00926922,
0.06616993, -0.09528868, 0.06353744, -0.10300278, -0.04216977,
0.13834757, -0.0439804 , -0.05489451, 0.05748696, -0.02646153,
0.02639312, -0.1424805 , -0.05328764, -0.11360066, -0.05215862,
0.02328033, 0.06903777, -0.03831793, -0.0649631 , -0.02677068,
0.07705265, 0.11282316, -0.06039653, 0.08879586, 0.02900434,
0.03495867, 0.07618292, -0.15206559, -0.06996718, -0.10180831,
-0.01852681, 0.0621195 , -0.08078951, -0.06472867, 0.00649193,
-0.02906965, -0.00562615, -0.10181142, 0.14631483, -0.02691234,
-0.0449581 , -0.06768148, -0.14837843, 0.00591947, 0.13189799,
0.08950461, -0.09030728, -0.03431619, -0.15363187, 0.10463613,
-0.06360365, 0.03076407, -0.23843719, 0.12627779, 0.00943171,
-0.03152905, -0.04594371, -0.01427775, 0.02384799, 0.00763995,
-0.01302205, 0.01930667, 0.19256516, 0.04888356, -0.02315179,
-0.18147527, -0.02883495, -0.24427226, -0.05883946, 0.09711844,
0.02236536, 0.16532755, -0.05398215, -0.08104754, 0.00680975,
-0.2076412 , 0.19293354, 0.02048309, -0.14072204, -0.06431156,
-0.06882941, 0.10384997, -0.01201017, -0.06934526, -0.02065195,
-0.15377323, 0.02488887, 0.01642702, -0.11942345, 0.03666817,
-0.04260147, 0.0460966 , 0.19208308, -0.00149917, 0.09103897,
-0.08409246, -0.00229917, 0.0308649 , -0.09187246, -0.13771881,
0.02569543, 0.05528186, 0.01997439, 0.0068558 , 0.05850806,
0.13373522, 0.06533468, -0.11358334, -0.05569442, -0.13252501,
0.0317649 , 0.06107381, -0.04135463, 0.09990298, 0.1721155 ,
-0.0232862 , -0.00518754, -0.08392647, 0.10745225, -0.04787033,
0.07003915, 0.04473805, -0.05343888, -0.07590986, 0.09890503,
-0.13220763, 0.15761884, 0.01254966, 0.07159693, 0.2163922 ,
0.0706775 , -0.08037703, -0.02174817, 0.00922287, -0.14820273,
0.07418934, 0.05675528, 0.05901892, 0.08499168, -0.07088676,
0.04398298, -0.01116003, 0.07408264, 0.03731104, -0.09286404,
-0.05726976, -0.09286391, -0.06280293, 0.07435899, -0.05318311,
-0.08465645, -0.07101735, -0.09878317, -0.02315412, 0.04980372,
0.03120912, 0.06119568, 0.08196454, -0.01675618, 0.10395606,
0.07163345, 0.08211765, -0.03836129, 0.08776304, 0.05949511,
0.01261996, 0.07629557, -0.03077972, 0.1014216 , 0.02663247,
-0.25162402, 0.05324515, -0.17487867, 0.02626908, -0.01785631,
-0.01677652, 0.06310172, 0.11175925, 0.13500917, 0.02175732,
0.13348427, -0.04852588, -0.15018572, -0.01485547, 0.09306358,
0.07984833, 0.06227739, 0.08456677, -0.1992962 , -0.02070233,
-0.12772235, 0.03287375, 0.00196922, 0.09598093, -0.06379709,
-0.10833501, 0.05440256, -0.12777323, 0.00854843, -0.01612958,
0.03918738, 0.08117849, -0.14819548, 0.02658651, 0.09616554,
-0.00249762, 0.10455485, 0.06745086, -0.04041155, 0.04394639,
0.02677601, 0.1049852 , 0.01489662, 0.1538879 , 0.07849001,
0.06335338, -0.10787047, -0.01897711, -0.08735528, -0.09845153,
-0.04195962, -0.02259701, -0.07576959, -0.02728209, -0.02308704,
0.04064317, -0.09514683, 0.12956946, 0.02450153, -0.04147912,
-0.07329746, -0.268567 , -0.04270407, -0.02746491, -0.08409931,
0.19540311, -0.00489267, 0.12485956, 0.00570589, 0.07255039,
0.00758378, -0.09491847, -0.1212426 , -0.08841562, -0.04497168,
-0.06164463, 0.03163375, -0.00016094, 0.2162991 , -0.0706714 ,
0.0260719 , -0.07955409, -0.13185492, 0.05533328, -0.01741955,
-0.09893776, 0.13881136, -0.05906575, 0.00042838, 0.10324635,
0.05969812, 0.1739419 , 0.00114153, -0.08487915, 0.02052278,
0.00210753, -0.09841961, -0.07115474, 0.05895515, 0.07927404,
-0.07059009, 0.10411056, -0.03169583, 0.09279449, -0.03314509,
-0.07824375, -0.10872853, -0.07618266, 0.02892239, 0.1517952 ,
-0.02554395, 0.0085359 , -0.06679222, 0.09961466, 0.03327283],
dtype=float32)
Using t-SNE we can reduce the dimensionality of each word’s vector from 300 to only two. This enables us to plot the points and learn how their position on the plot and the distances between each other corresponds to their meaning:
# reduce vectors to two dimensions
from sklearn.manifold import TSNE
tsne = TSNE(n_components=2, random_state=1)
emb_val = list(words_embeddings.values())
emb_name = list(words_embeddings.keys())
X_2d = tsne.fit_transform(emb_val)
tsne_df = pd.DataFrame({"X": X_2d[:, 0], "Y": X_2d[:, 1], "word": emb_name})
tsne_df.head(3)
| X | Y | word | |
|---|---|---|---|
| 0 | 11.538602 | -7.251408 | und |
| 1 | 32.283974 | -20.078789 | ich |
| 2 | 34.469269 | -19.970659 | die |
# plotting the reduced embeddingd vectors
from hvplot import pandas
import holoviews as hv
hv.extension("bokeh")
hv.Scatter(tsne_df.sample(1000), vdims=["Y", "word"], kdims=["X"]).opts(
size=7, tools=["hover"], width=550, height=550, title="Visualizing Embeddings"
)
What can we learn from the plot? Words in the upper left corner seem to all be very positiv. They probably are good in predicting good ratings. Words with a low Y value seem to be more negative in general. At around X=30 and Y=20 there is a cluster of words which are all surnames of doctors. Our neural network has learned to identify names from the comments even though this was not a direct objective of the model. This is astonishing and an exciting side effect of embeddings: words with similar meanings might be represented by similar vectors. This is exactly what happend here.
We can also calculate which word vectors are very close to each other by using the KDTree algorithm. Then, we look for words that are most similar to “schlecht” (bad):
from sklearn.neighbors import KDTree
tree = KDTree(tsne_df[["X", "Y"]])
# get most similar vectors
nearest = tree.query(tsne_df.loc[tsne_df["word"] == "schlecht", ["X", "Y"]], k=5)
tsne_df.iloc[nearest[1][0], :]
| X | Y | word | |
|---|---|---|---|
| 510 | -13.225616 | -46.543968 | schlecht |
| 4876 | -12.826276 | -46.083984 | fremdwort |
| 2204 | -12.770123 | -46.042770 | frech |
| 5496 | -12.951906 | -45.229324 | geweint |
| 8708 | -12.083499 | -45.492523 | kälte |
It seems that vectors close to the vector of “schlecht” also hold a negative meaning. For example, we find “frech” (audacious) and “geweint” (whining). What about positive words? We look for the vectors close to “gut” (good):
# get most similar vectors
nearest = tree.query(tsne_df.loc[tsne_df["word"] == "gut", ["X", "Y"]], k=5)
tsne_df.iloc[nearest[1][0], :]
| X | Y | word | |
|---|---|---|---|
| 28 | -32.525509 | 42.271885 | gut |
| 2455 | -32.821663 | 42.302612 | sanft |
| 404 | -32.472672 | 41.609344 | positiv |
| 2770 | -31.886206 | 42.486290 | beeindruckend |
| 3728 | -31.645645 | 41.917389 | engel |
Again, our expectation is confirmed: We find “sanft” (gentle) and “engel” (angle) and more fitting words.
Evaluation
Finally, we’re excited to see how well our model performs. Recall that the best model so far (from part1) achieved a macro f1 score of 0.90:
# Load best model from Checkpoint
# model = load_model(PATH_CHECKPOINTS+"lstm-no-embed/Model_300emb_06_0.0757.h5",
# compile=False)
# Predict on test data
pred = model.predict(X_test_vec)
pred_class = (pred > 0.5).astype(int)
pred_len = X_test_vec.shape[0]
report = metrics.classification_report(Y_test, pred_class[0:pred_len])
print(report)
precision recall f1-score support
0.0 0.99 0.98 0.99 76290
1.0 0.84 0.89 0.87 8305
accuracy 0.97 84595
macro avg 0.92 0.94 0.93 84595
weighted avg 0.97 0.97 0.97 84595
We increase our prediction and achieve a macro f1 score of 0.93. The best part about this is that we are able significantly increase our prediction performance of class 1 (negative ratings). This class is way more challenging to predict. Still, we increased our recall from 0.79 to 0.89 and the f1-score from 0.82 to 0.87. So using this more advanced approach has been a great success! There still are a lot of parts and pieces that could be tweaked in this model. This might further increase our scores. Basically, all parameters defined at the top of the notebook could be changed and evaluated. Moreover, we could change the architecture of the neural network and / or make it more complex / deep. Possibilities are unlimited. However, the setup presented here should already be pretty decent.
Using Pre-trained embeddings
Instead of creating embeddings from scratch as part of a neural network for a specific task, it is also possible to use pre trained embeddings. These have been generated using huge models and data sets and contain large dictionaries. Usually, the vectors of pre trained embeddings capture semantic meaning. For example, two words that are semantic substitues will also have very similar vectors.
Popular choices for embeddings include Word2Vec, GloVe and FastText.
In order to try to improve our neural network model architecture, we will use FastText embeddings. We use the German FastText embeddings that have been trained on CommonCrawl and German Wikipedia. Let’ download and extract the vectors:
!wget "https://dl.fbaipublicfiles.com/fasttext/vectors-crawl/cc.de.300.vec.gz"
!gzip -d cc.de.300.vec.gz
--2020-06-07 17:42:47-- https://dl.fbaipublicfiles.com/fasttext/vectors-crawl/cc.de.300.vec.gz
Resolving dl.fbaipublicfiles.com (dl.fbaipublicfiles.com)... 172.67.9.4, 104.22.74.142, 104.22.75.142, ...
Connecting to dl.fbaipublicfiles.com (dl.fbaipublicfiles.com)|172.67.9.4|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 1278030050 (1.2G) [binary/octet-stream]
Saving to: ‘cc.de.300.vec.gz’
cc.de.300.vec.gz 100%[===================>] 1.19G 25.2MB/s in 49s
2020-06-07 17:43:36 (24.8 MB/s) - ‘cc.de.300.vec.gz’ saved [1278030050/1278030050]
An easy way to use the vectors is the gensim library:
%%time
# Load Fasttext vector embeddings - takes some time 10min
# use pickle to dump loaded model -> load that = 12s
from gensim.models import KeyedVectors
de_model = KeyedVectors.load_word2vec_format(PATH_DATA + "/cc.de.300.vec")
# pickle.dump(de_model, open(PATH_TMP+"/de_model.pkl", "wb"))
# de_model = pickle.load(open(PATH_TMP+"/de_model.pkl", "rb"))
CPU times: user 8min 53s, sys: 8.44 s, total: 9min 1s
Wall time: 8min 54s
I’ve mentioned before that similar words are represented by similar vectors. Let’s look at the work “arzt” (male doctor):
de_model.similar_by_word("arzt")[0:5]
[('hausarzt', 0.8440093994140625),
('artzt', 0.8014777302742004),
('arzt.', 0.7992661595344543),
('kinderarzt', 0.7868670225143433),
('frauenarzt', 0.7781710624694824)]
Another astonishing quality of the embeddings is that vector calculations represent semantic relationships. Let’s try this out:
# get vectors for the words
woman = de_model.get_vector("frau")
doctor = de_model.get_vector("arzt")
# calculate and get closest match for resulting vector
de_model.similar_by_vector(doctor + woman)
[('arzt', 0.8891544938087463),
('frau', 0.7991741895675659),
('frauenarzt', 0.7820956707000732),
('hausarzt', 0.7763659954071045),
('ärztin', 0.7679202556610107),
('frauenärztin', 0.7507506608963013),
('arzthelferin', 0.7356253862380981),
('kinderärztin', 0.7130227088928223),
('hausärztin', 0.7106521129608154),
('kinderarzt', 0.7027797698974609)]
Wee add the vector of woman to the vector of doctor. Then, we look for words whose vector is close to the resulting vector. Remarkably, amongst the results we find “ärztin” (female doctor) and “frauenarzt” (gynaecologist)!
Preparing the embeddings
Now, let’s see if these properties will help out with our model performance. To get started, we need to assign the correct vector to each of the unique tokens in our comment texts:
# Create embedding matrix
print("preparing embedding matrix...")
words_not_found = []
# all words from the comments
word_index = tokenizer.word_index
# max unique words to keep
nb_words = min(MAX_FEATURES, len(word_index))
# define matrix dimensions
embedding_matrix = np.zeros((nb_words, EMBED_DIM))
for word, i in word_index.items():
if i >= nb_words:
continue
try:
embedding_vector = de_model.get_vector(word)
except KeyError:
embedding_vector = None
if (embedding_vector is not None) and len(embedding_vector) > 0:
# words not found in embedding index will be all-zeros.
embedding_matrix[i] = embedding_vector
else:
words_not_found.append(word)
print(f"Null word embeddings: {np.sum(np.sum(embedding_matrix, axis=1) == 0)}")
print(
f"Some of the words not found:\n"
f"{' '.join([random.choice(words_not_found) for x in range(0,10)])}"
)
preparing embedding matrix...
Null word embeddings: 8750
Some of the words not found:
behrendt knorpelschadens giers kuhlmann bärenklau umfeldes platzer besenreisern rimbach servicepersonal
While the FastText embeddings include a lot of words, some of the words in our comments are still missing. However, its only a small fraction and you can see that those are mostly surnames or very special medical terms. Hence, we can ignore this for now. We can now use this embedding matrix in our neural network. We use it to specify the weights of the embedding layer:
# Define NN architecture
with strategy.scope():
model = Sequential()
model.add(
Embedding(
input_dim=MAX_FEATURES,
output_dim=EMBED_DIM,
input_length=MAXLEN,
weights=[embedding_matrix],
trainable=False,
)
)
model.add(Dropout(0.3))
model.add(
Conv1D(FILTERS, KERNEL_SIZE, padding="valid", activation="relu", strides=1)
)
model.add(MaxPooling1D(pool_size=POOL_SIZE))
model.add(LSTM(LSTM_OUTPUT_SIZE))
model.add(Dense(1, activation="sigmoid"))
model.compile(
loss="binary_crossentropy",
optimizer=tf.keras.optimizers.RMSprop(),
metrics=["accuracy"],
)
model.summary()
Model: "sequential"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
embedding (Embedding) (None, 355, 300) 9000000
_________________________________________________________________
dropout (Dropout) (None, 355, 300) 0
_________________________________________________________________
conv1d (Conv1D) (None, 351, 64) 96064
_________________________________________________________________
max_pooling1d (MaxPooling1D) (None, 87, 64) 0
_________________________________________________________________
lstm (LSTM) (None, 100) 66000
_________________________________________________________________
dense (Dense) (None, 1) 101
=================================================================
Total params: 9,162,165
Trainable params: 162,165
Non-trainable params: 9,000,000
_________________________________________________________________
Because we specified the weights and set trainable = False for the embedding layer the model won’t need to learn new embeddings but just use the ones we provided. Hence, the number of trainable parameters of the model is greatly reduced compared to our first model.
That was all we needed to change, so let’s fit this new model:
%%time
# Stop training when validation acc starts dropping
# Save checkpoint of model each period
now = datetime.now().strftime("%Y-%m-%d_%H%M")
# Create callbacks
callbacks = [
EarlyStopping(monitor="val_loss", verbose=1, patience=2),
ModelCheckpoint(
PATH_CHECKPOINTS + now + "_Model_FT-Embed_{epoch:02d}_{val_loss:.4f}.h5",
monitor="val_loss",
save_best_only=True,
verbose=1,
),
]
# Fit the model
steps_per_epoch = int(np.floor((len(X_train_vec) / BATCH_SIZE)))
print(
f"Model Params.\nbatch_size: {BATCH_SIZE}\nEpochs: {EPOCHS}\n"
f"Step p. Epoch: {steps_per_epoch}\n"
)
hist = model.fit(
X_train_vec,
Y_train,
batch_size=BATCH_SIZE,
epochs=EPOCHS,
steps_per_epoch=steps_per_epoch,
callbacks=callbacks,
validation_data=(X_test_vec, Y_test),
)
Model Params.
batch_size: 256
Epochs: 10
Step p. Epoch: 991
Epoch 1/10
989/991 [============================>.] - ETA: 0s - accuracy: 0.9196 - loss: 0.1989
Epoch 00001: val_loss improved from inf to 0.12653, saving model to /drive/My Drive/tmp/2020-06-07_1904_Model_FT-Embed_01_0.1265.h5
991/991 [==============================] - 22s 22ms/step - accuracy: 0.9197 - loss: 0.1987 - val_accuracy: 0.9471 - val_loss: 0.1265
Epoch 2/10
990/991 [============================>.] - ETA: 0s - accuracy: 0.9434 - loss: 0.1322
Epoch 00002: val_loss improved from 0.12653 to 0.10639, saving model to /drive/My Drive/tmp/2020-06-07_1904_Model_FT-Embed_02_0.1064.h5
991/991 [==============================] - 20s 20ms/step - accuracy: 0.9434 - loss: 0.1322 - val_accuracy: 0.9571 - val_loss: 0.1064
Epoch 3/10
991/991 [==============================] - ETA: 0s - accuracy: 0.9524 - loss: 0.1146
Epoch 00003: val_loss did not improve from 0.10639
991/991 [==============================] - 20s 20ms/step - accuracy: 0.9524 - loss: 0.1146 - val_accuracy: 0.9577 - val_loss: 0.1098
Epoch 4/10
988/991 [============================>.] - ETA: 0s - accuracy: 0.9576 - loss: 0.1046
Epoch 00004: val_loss improved from 0.10639 to 0.09166, saving model to /drive/My Drive/tmp/2020-06-07_1904_Model_FT-Embed_04_0.0917.h5
991/991 [==============================] - 20s 20ms/step - accuracy: 0.9576 - loss: 0.1045 - val_accuracy: 0.9637 - val_loss: 0.0917
Epoch 5/10
991/991 [==============================] - ETA: 0s - accuracy: 0.9585 - loss: 0.1023
Epoch 00005: val_loss did not improve from 0.09166
991/991 [==============================] - 20s 20ms/step - accuracy: 0.9585 - loss: 0.1023 - val_accuracy: 0.9566 - val_loss: 0.0985
Epoch 6/10
989/991 [============================>.] - ETA: 0s - accuracy: 0.9598 - loss: 0.0979
Epoch 00006: val_loss improved from 0.09166 to 0.08998, saving model to /drive/My Drive/tmp/2020-06-07_1904_Model_FT-Embed_06_0.0900.h5
991/991 [==============================] - 20s 20ms/step - accuracy: 0.9598 - loss: 0.0979 - val_accuracy: 0.9575 - val_loss: 0.0900
Epoch 7/10
990/991 [============================>.] - ETA: 0s - accuracy: 0.9623 - loss: 0.0908
Epoch 00007: val_loss improved from 0.08998 to 0.08407, saving model to /drive/My Drive/tmp/2020-06-07_1904_Model_FT-Embed_07_0.0841.h5
991/991 [==============================] - 20s 20ms/step - accuracy: 0.9622 - loss: 0.0908 - val_accuracy: 0.9670 - val_loss: 0.0841
Epoch 8/10
989/991 [============================>.] - ETA: 0s - accuracy: 0.9637 - loss: 0.0889
Epoch 00008: val_loss improved from 0.08407 to 0.08192, saving model to /drive/My Drive/tmp/2020-06-07_1904_Model_FT-Embed_08_0.0819.h5
991/991 [==============================] - 20s 20ms/step - accuracy: 0.9637 - loss: 0.0889 - val_accuracy: 0.9675 - val_loss: 0.0819
Epoch 9/10
991/991 [==============================] - ETA: 0s - accuracy: 0.9641 - loss: 0.0872
Epoch 00009: val_loss improved from 0.08192 to 0.08085, saving model to /drive/My Drive/tmp/2020-06-07_1904_Model_FT-Embed_09_0.0809.h5
991/991 [==============================] - 21s 21ms/step - accuracy: 0.9641 - loss: 0.0872 - val_accuracy: 0.9674 - val_loss: 0.0809
Epoch 10/10
991/991 [==============================] - ETA: 0s - accuracy: 0.9658 - loss: 0.0828
Epoch 00010: val_loss did not improve from 0.08085
991/991 [==============================] - 19s 20ms/step - accuracy: 0.9658 - loss: 0.0828 - val_accuracy: 0.9652 - val_loss: 0.0872
CPU times: user 38.3 s, sys: 8.47 s, total: 46.7 s
Wall time: 3min 31s
We can now check the prediction results:
# Predict on test data
pred = model.predict(X_test_vec)
pred_class = (pred > 0.5).astype(int)
pred_len = X_test_vec.shape[0]
report = metrics.classification_report(Y_test, pred_class[0:pred_len])
print(report)
precision recall f1-score support
0.0 0.97 0.99 0.98 76330
1.0 0.88 0.74 0.81 8265
accuracy 0.97 84595
macro avg 0.93 0.87 0.89 84595
weighted avg 0.96 0.97 0.96 84595
The resulting model performs decent, but significantly worse than our original LSTM neural network. We achieve a macro f1-score of only 0.89. The precision of class 1 increases, but at the same time the recall drops significantly. However, because we use embeddings that were trained for a much more general task but not for binary text classification, this is not very surprising. Also, we can easily work around this limitation by allowing our model to further train the embeddings. This is the concept of transfer learning: we use the weights from a model pre trained on a similar, but more general task as starting point. Then, we allow our model to further refine those weights for our specific task. This strategy is extremly powerful, especially in cases where you have only limited data for your specific task. I’m wondering how good it will perform here:
# Define NN architecture
with strategy.scope():
model = Sequential()
model.add(
Embedding(
input_dim=MAX_FEATURES,
output_dim=EMBED_DIM,
input_length=MAXLEN,
weights=[embedding_matrix],
trainable=True,
)
)
model.add(Dropout(0.3))
model.add(
Conv1D(FILTERS, KERNEL_SIZE, padding="valid", activation="relu", strides=1)
)
model.add(MaxPooling1D(pool_size=POOL_SIZE))
model.add(LSTM(LSTM_OUTPUT_SIZE))
model.add(Dense(1, activation="sigmoid"))
model.compile(
loss="binary_crossentropy",
optimizer=tf.keras.optimizers.RMSprop(),
metrics=["accuracy"],
)
model.summary()
Model: "sequential_1"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
embedding_1 (Embedding) (None, 355, 300) 9000000
_________________________________________________________________
dropout_1 (Dropout) (None, 355, 300) 0
_________________________________________________________________
conv1d_1 (Conv1D) (None, 351, 64) 96064
_________________________________________________________________
max_pooling1d_1 (MaxPooling1 (None, 87, 64) 0
_________________________________________________________________
lstm_1 (LSTM) (None, 100) 66000
_________________________________________________________________
dense_1 (Dense) (None, 1) 101
=================================================================
Total params: 9,162,165
Trainable params: 9,162,165
Non-trainable params: 0
_________________________________________________________________
%%time
# Stop training when validation acc starts dropping
# Save checkpoint of model each period
now = datetime.now().strftime("%Y-%m-%d_%H%M")
# Create callbacks
callbacks = [
EarlyStopping(monitor="val_loss", verbose=1, patience=2),
ModelCheckpoint(
PATH_CHECKPOINTS
+ now
+ "_Model_FT-Embed-trainable_{epoch:02d}_{val_loss:.4f}.h5",
monitor="val_loss",
save_best_only=True,
verbose=1,
),
]
# Fit the model
steps_per_epoch = int(np.floor((len(X_train_vec) / BATCH_SIZE)))
print(
f"Model Params.\nbatch_size: {BATCH_SIZE}\nEpochs: {EPOCHS}\n"
f"Step p. Epoch: {steps_per_epoch}\n"
)
hist = model.fit(
X_train_vec,
Y_train,
batch_size=BATCH_SIZE,
epochs=EPOCHS,
steps_per_epoch=steps_per_epoch,
callbacks=callbacks,
validation_data=(X_test_vec, Y_test),
)
Model Params.
batch_size: 256
Epochs: 10
Step p. Epoch: 991
Epoch 1/10
991/991 [==============================] - ETA: 0s - accuracy: 0.9414 - loss: 0.1579
Epoch 00001: val_loss improved from inf to 0.12406, saving model to /drive/My Drive/tmp/2020-06-07_1914_Model_FT-Embed-trainable_01_0.1241.h5
991/991 [==============================] - 27s 27ms/step - accuracy: 0.9414 - loss: 0.1579 - val_accuracy: 0.9500 - val_loss: 0.1241
Epoch 2/10
990/991 [============================>.] - ETA: 0s - accuracy: 0.9636 - loss: 0.0986
Epoch 00002: val_loss improved from 0.12406 to 0.09523, saving model to /drive/My Drive/tmp/2020-06-07_1914_Model_FT-Embed-trainable_02_0.0952.h5
991/991 [==============================] - 25s 26ms/step - accuracy: 0.9636 - loss: 0.0986 - val_accuracy: 0.9651 - val_loss: 0.0952
Epoch 3/10
990/991 [============================>.] - ETA: 0s - accuracy: 0.9647 - loss: 0.0913
Epoch 00003: val_loss did not improve from 0.09523
991/991 [==============================] - 25s 25ms/step - accuracy: 0.9648 - loss: 0.0913 - val_accuracy: 0.9618 - val_loss: 0.0987
Epoch 4/10
991/991 [==============================] - ETA: 0s - accuracy: 0.9732 - loss: 0.0699
Epoch 00004: val_loss improved from 0.09523 to 0.07889, saving model to /drive/My Drive/tmp/2020-06-07_1914_Model_FT-Embed-trainable_04_0.0789.h5
991/991 [==============================] - 25s 25ms/step - accuracy: 0.9732 - loss: 0.0699 - val_accuracy: 0.9693 - val_loss: 0.0789
Epoch 5/10
991/991 [==============================] - ETA: 0s - accuracy: 0.9781 - loss: 0.0579
Epoch 00005: val_loss improved from 0.07889 to 0.06569, saving model to /drive/My Drive/tmp/2020-06-07_1914_Model_FT-Embed-trainable_05_0.0657.h5
991/991 [==============================] - 25s 25ms/step - accuracy: 0.9781 - loss: 0.0579 - val_accuracy: 0.9743 - val_loss: 0.0657
Epoch 6/10
990/991 [============================>.] - ETA: 0s - accuracy: 0.9814 - loss: 0.0506
Epoch 00006: val_loss did not improve from 0.06569
991/991 [==============================] - 25s 25ms/step - accuracy: 0.9814 - loss: 0.0506 - val_accuracy: 0.9734 - val_loss: 0.0722
Epoch 7/10
991/991 [==============================] - ETA: 0s - accuracy: 0.9833 - loss: 0.0454
Epoch 00007: val_loss did not improve from 0.06569
991/991 [==============================] - 25s 25ms/step - accuracy: 0.9833 - loss: 0.0454 - val_accuracy: 0.9748 - val_loss: 0.0698
Epoch 00007: early stopping
CPU times: user 29.3 s, sys: 6.73 s, total: 36 s
Wall time: 3min 8s
After allowing our embedding layer to further refine the pre trained FastText embeddings, here is the result:
# Predict on test data
pred = model.predict(X_test_vec)
pred_class = (pred > 0.5).astype(int)
pred_len = X_test_vec.shape[0]
# Show prediction metrics
report = metrics.classification_report(Y_test, pred_class[0:pred_len])
print(report)
precision recall f1-score support
0.0 0.99 0.98 0.99 76330
1.0 0.86 0.89 0.87 8265
accuracy 0.97 84595
macro avg 0.92 0.94 0.93 84595
weighted avg 0.98 0.97 0.97 84595
On the first look, this model performs way better than the previous one and as good as our first model. It has a macro f1-score of 0.93. However, a more thorough examination reveals that this model is a slight improvement. For class 1, we increased the precision from 0.84 to 0.86 while keeping the recall and class 0 metrics constant. So using a LTSM neural network with fine tuned FastText embeddings is our most performant classifier so far!
Conclusion
In this second part of our series, we have advanced our binary text classification model of German comments of patients on their doctors. For that, we have implemented different LTSM neural network architectures using Keras. Further, we have gained some understanding on word embeddings and learned about their properties by inspecting and visualizing the embedding vectors calculated for our data. Finally, we have introduced pre trained embeddings using FastText and further fine tuned them. As a result, we were able to create a very powerful prediction model. The classifier is able to assign the correct sentiment to comments and predict their raitings with high proficiency.
In the next part of this series, we will continue our NLP journey. We will introduce state of the art models (i.e. transformer neural networks like BERT) to further push the boundaries of our classifier.